Abstract
Introduction
Genetic abnormalities, such as MYC oncogene rearrangements, contribute to the outcome heterogeneity in diffuse large B-cell lymphoma (DLBCL) patients. These rearrangements occur in 10-15% of DLBCL patients and have been associated with a poor prognosis. Recently, radiomics features extracted from PET/CT scans have shown to be predictive of outcome. The aim of this study was to investigate if the ability to predict outcome in DLBCL can be improved by combining different clinical, radiomics and genetic features.
Methods
323 DLBCL patients from the HOVON-84, HOVON-130, and PETAL trials with a baseline PET/CT scan and a minimum follow-up of two years were included. MYC status was assessed using Fluorescence in situ hybridization (FISH). 245 patients were MYC negative, whereas 25 patients had a MYC rearrangement and 57 patients had MYC and BCL2 and/or BCL6 rearrangements. Lesions were delineated using a semi-automated preselection of 18F-FDG avid structures defined by a SUV4.0 threshold using the ACCURATE tool. Next, 5 conventional PET features (maximum standardized uptake value (SUV max), SUV peak, SUV mean, metabolic tumor volume (MTV) and total lesion glycolysis and 18 dissemination features were extracted. Dissemination features were pertaining to distance between lesions, differences in uptake between lesions and differences in volume between lesions. Logistic regression with backward feature selection was used to predict 2-year time to progression, defined as time from baseline PET/CT to progression. We tested the predictive value of 4 models. 1) a clinical model using individual components of the international prognostic index (IPI): Ann Arbor stage (categorical), WHO performance status (categorical), lactate dehydrogenase (LDH) levels (dichotomous) and age (continuous), 2) a model that included clinical and genetic predictors: MYC status (categorical) and IPI components, 3) a model that included radiomics features: 5 conventional PET and 18 dissemination features and 4) a model that combined clinical and genetic predictors with radiomics features. Model performance was assessed using repeated cross-validation (5-fold, 1000 repeats) yielding the cross-validated area under the curve of the receiver-operator-characteristics curve (CV-AUC). To match prevalence of MYC-positive patients with real-world prevalence (Rosenwald et al, JCO 2019) all 245 MYC-negative patients were used for each repeat, and 10 MYC-FISH_positive DLBCL patients and 20 patients with MYC and BCL2 and/or BCL6 rearrangements were selected using random stratified sampling. Regression coefficients were averaged over all folds to calculate the probability of progression for all patients. High- and low-risk groups were defined based on prevalence of events and the diagnostic performance was assessed using positive- and negative predictive values.
Results
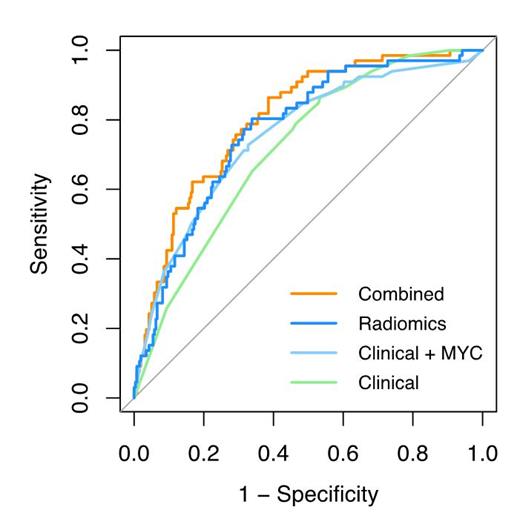
The highest model performance for the clinical model was observed when combining Ann Arbor stage, LDH and extranodal involvement and yielded in a CV-AUC of 0.69 (95% confidence interval (CI): 0.52-0.83). MYC status combined with WHO performance status, LDH and extranodal involvement yielded an improved CV-AUC of 0.71 (95% CI: 0.52-0.86). The highest model performance for the radiomics model was observed for MTV combined with the maximum distance between the largest lesion and any other lesions (Dmax bulk), the maximum difference in SUV peak between two lesions (DSUVpeak patient) and the maximum difference in volume between two lesions (DVol patient) yielding a CV-AUC of 0.77 (95% CI: 0.62-0.90). The optimal combined model included MYC status, WHO performance status, LDH, MTV, Dmax patient, DSUVpeak patient and DVol patient after backward feature selection and yielded a CV-AUC of 0.77 (95% CI: 0.60 - 0.90). The positive predictive value was highest for the combined model (53.0%) and increased by 20% compared to the optimal clinical model (33.1%). Negative predictive values were comparable between models and ranged between 85.8-88.1%.
Conclusions
Prediction models using 18F-FDG PET/CT radiomics features at baseline aids in identifying DLBCL patients at high risk for relapse. The positive predictive value increased when radiomics features were added to the clinical and genetic parameters. Therefore, radiomics features can increase the efficiency of clinical trials by only selecting poor prognosis patients.
Chamuleau: Gilead: Research Funding; Genmab: Research Funding; Celgene: Research Funding. Lugtenburg: Incyte: Honoraria; Regeneron: Honoraria; Genmab: Honoraria; Servier: Honoraria, Research Funding; Takeda: Honoraria, Research Funding; Celgene: Honoraria, Other: Non-financial support; Travel support; Roche: Honoraria, Research Funding. Dührsen: CPT Cellex Patient Treatment: Honoraria, Membership on an entity's Board of Directors or advisory committees; Amgen: Honoraria, Membership on an entity's Board of Directors or advisory committees. Hüttmann: Celgene: Honoraria; Gilead: Honoraria; Lead Discovery Center GmbH: Consultancy; Takeda: Honoraria, Membership on an entity's Board of Directors or advisory committees. Richter: HTG Molecular Diagnostics, Inc.: Current Employment, Research Funding. Klapper: Regeneron: Consultancy, Research Funding; Amgen: Research Funding; Roche: Consultancy, Research Funding; Takeda: Consultancy, Research Funding. Zijlstra: Takeda: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal